Replaying
from py_replay_bg.replay.forcing_ra import CustomRaBase--- sidebar: heading
Replaying
Once created, we are finally ready to use the digital twins to run replay simulations.
The aims can be A LOT, e.g.:
- you want to simulate the impact of modulating the input data you used for twinning, e.g., reducing the insulin bolus by 30%, and see what happens to glucose control
- you want to evaluate the performance of a bolus calculator you developed
- you want to develop a new algorithm for type 1 diabetes management
- you want to find the best hypotreatment policy able to prevent hypoglycemia
- you want to test an artificial pancreas algorithm
- you just want to generate data using random insulin/meal inputs
- and so on and so forth...
In the following, we present how to use ReplayBG for running replay simulations and we will provide a comprehensive list of its capabilities.
The replay method
Replay simulations in ReplayBG can be performed using the replay method of the ReplayBG object, which is formally defined as:
rbg.replay(data: pd.DataFrame,
bw: float,
save_name: str,
x0: np.ndarray | None = None,
previous_data_name: str | None = None,
twinning_method: str = 'mcmc',
bolus_source: str = 'data',
basal_source: str = 'data',
cho_source: str = 'data',
meal_generator_handler: Callable = default_meal_generator_handler,
meal_generator_handler_params: Dict | None = None,
bolus_calculator_handler: Callable = standard_bolus_calculator_handler,
bolus_calculator_handler_params: Dict | None = None,
basal_handler: Callable = default_basal_handler,
basal_handler_params: Dict | None = None,
enable_hypotreatments: bool = False,
hypotreatments_handler: Callable = ada_hypotreatments_handler,
hypotreatments_handler_params: Dict | None = None,
enable_correction_boluses: bool = False,
correction_boluses_handler: Callable = corrects_above_250_handler,
correction_boluses_handler_params: Dict | None = None,
save_suffix: str = '',
save_workspace: bool = False,
n_replay: int = 1000,
sensors: list | None = None,
sensor_cgm: CGM = Vettoretti19CGM,
snack_absorption=snack_absorption,
snack_absorption_delay=snack_absorption_delay,
hypotreatment_absorption=hypotreatment_absorption,
custom_Ra: CustomRaBase = None
) -> Dict:Input parameters
data: A Pandas dataframe which contains the data to be used by the tool. For more information ondataformat and requirements see Data Requirements page.bw: A float representing the patient's body weight in kg.save_name: A string used to label, thus identify, each output file and result. This MUST correspond to thesave_namestring provided to thetwinmethod during twinning.x0, optional, default:None: An np.ndarray, containing the initial model conditions. IfNone, the model will start to the default steady state.previous_data_name, optional, default:None: A string representing the name of the previous data portion. This is used to correcly "transfer" the initial model conditions to the current portion of data. Practically, this is equal to thesave_nameused during the creation of the digital twin related to the previous portion of data. It must be set ifx0is notNone.twinning_method, optional,{'mcmc', 'map'}, default:'mcmc': A string used to select the method to be used to twin the model. This MUST correspond to thetwinning_methodvalue provided to thetwinmethod during twinning.bolus_source, optional, {'data','dss'}, default:'data': A string defining whether to use, during replay, the insulin bolus data contained in the'data'dataframe (if'data'), or the boluses generated by the bolus calculator implemented via the provided'bolusCalculatorHandler'function.basal_source, optional, {'data','u2ss','dss'}, default:'data': A string defining whether to use,- during replay, the insulin basal data contained in the
'data'dataframe (if'data'), or the basal generated by the controller implemented via the provided'basalControllerHandler'function (if'dss'), or fixed to the average basal rate used during twinning (if'u2ss'). cho_source, optional, {'data','generated'}, default:'data': A string defining whether to use, during replay, the CHO data contained in the'data'dataframe (if'data'), or the CHO generated by the meal generator- implemented via the provided
'mealGeneratorHandler'function (if'generated'). meal_generator_handler, optional, default:default_meal_generator_handler: A callback function that implements a meal generator to be used during the replay simulation. For more information see the below Event handlers section.meal_generator_handler_params, optional, default:None: A Python dictionary that contains the parameters to pass to themeal_generator_handlerfunction. It also serves as memory area for themeal_generator_handlerfunction. For more information see the below Event handlers section.bolus_calculator_handler, optional, default :standard_bolus_calculator_handler: A callback function that implements a bolus calculator to be used during the replay simulation. For more information see the below Event handlers section.bolus_calculator_handler_params, optional, default:None: A Python dictionary that contains the parameters to pass to thebolusCalculatorHandlerfunction. It also serves as memory area for thebolusCalculatorHandlerfunction. For more information see the below Event handlers section.basal_handler, optional, default:default_basal_handler: A callback function that implements a basal controller to be used during the replay simulation. For more information see the below Event handlers section.basal_handler_params, optional, default:None: A Python dictionary that contains the parameters to pass to thebasalHandlerfunction. It also serves as memory area for thebasalHandlerfunction. For more information see the below Event handlers section.enable_hypotreatments, optional, default:False: A boolean that specifies whether to enable hypotreatments during the replay simulation. For more information see the below Event handlers section.hypotreatments_handler, optional, default:ada_hypotreatments_handler: A callback function that implements a hypotreatment strategy during the replay simulation. For more information see the below Event handlers section.hypotreatments_handler_params, optional, default:None: A Python dictionary that contains the parameters to pass to thehypoTreatmentsHandlerfunction. It also serves as memory area for thehypoTreatmentsHandlerfunction. For more information see the below Event handlers section.enable_correction_boluses, optional, default:False: A boolean that specifies whether to enable correction boluses during the replay simulation. For more information see the below Event handlers section.correction_boluses_handler, optional, default:corrects_above_250_handler: A callback function that implements a corrective bolus strategy during the replay simulation. For more information see the below Event handlers section.correction_boluses_handler_params, optional, default:None: A Python dictionary that contains the parameters to pass to thecorrectionBolusesHandlerfunction. It also serves as memory area for thecorrectionBolusesHandlerfunction. For more information see the below Event handlers section.save_suffix, optional, default:'': A string to be attached as suffix to the resulting output files' name.save_workspace, optional, default:False: A boolean that specifies whether to save the results of the simulation in theresults/workspacesfolder or not.n_replay, optional, {1, 10, 100, 1000}, default:1000: A number to select the sampled form to be used for the replay simulations. Ignored if twinning_method is 'map'.sensors: , optional, default:None: Alist[Sensors]to be used in each of the replay simulations. Its length must coincide with the selectedn_replay. Used when working with intervals. IfNonenew sensors will be used.sensor_cgm, optional, default:Vettoretti19CGM: The class of the CGM error model to be used during the replay simulation. For more information see the CGM Error Model page.snack_absorption, optional, default:None: A value to override the identified snack absorption rate.snack_absorption_delay, optional, default:None: A value to override the identified snack absorption delay (between 0 and 60 minutes)hypotreatment_absorption, optional, default:None: A value to override the identified hypotreatment absorption rate.custom_Ra, optional, default:None: An object that inherits fromCustomRaBaseand implements a custom glucose rate of appearance model to be used during the replay simulation. For more information see the Custom Ra Models page.
REMEMBER
The total length of the simulation, simulation_length, is defined in minutes and determined by ReplayBG automatically using the t column of data and the yts input parameter provided to the ReplayBG object builder.
For example, if yts is 5 minutes and t starts from 20-Dec-2013 10:36:00 and ends to 20-Dec-2013 10:46:00, then simulation_length will be 10.
Warning
The replay method will generate an error if the digital twin has not been created first.
Simulation results
The replay method will return the simulation results in the form of a dictionary with the following fields:
glucose: a dictionary containing the simulated plasma glucose timeseries (mg/dl). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing eachn_replaysimulated glucose profilemedian: a np.ndarray of size (simulation_length, ) containing the median simulated glucose profile (obtained fromrealizations)ci25th: a np.ndarray of size (simulation_length, ) containing the simulated glucose profile corresponding to the 25th percentile (obtained fromrealizations)ci75th: a np.ndarray of size (simulation_length, ) containing the simulated glucose profile corresponding to the 75th percentile (obtained fromrealizations)ci5th: a np.ndarray of size (simulation_length, ) containing the simulated glucose profile corresponding to the 5th percentile (obtained fromrealizations)ci95th: a np.ndarray of size (simulation_length, ) containing the simulated glucose profile corresponding to the 95th percentile (obtained fromrealizations)
cgm: a dictionary containing the simulated CGM timeseries (mg/dl). It contains:realizations: a np.ndarray of size (n_replay,simulation_length/yts) containing eachn_replaysimulated cgm profilemedian: a np.ndarray of size (simulation_length/yts, ) containing the median simulated cgm profile (obtained fromrealizations)ci25th: a np.ndarray of size (simulation_length/yts, ) containing the simulated cgm profile corresponding to the 25th percentile (obtained fromrealizations)ci75th: a np.ndarray of size (simulation_length/yts, ) containing the simulated cgm profile corresponding to the 75th percentile (obtained fromrealizations)ci5th: a np.ndarray of size (simulation_length/yts, ) containing the simulated cgm profile corresponding to the 5th percentile (obtained fromrealizations)ci95th: a np.ndarray of size (simulation_length/yts, ) containing the simulated cgm profile corresponding to the 95th percentile (obtained fromrealizations)
x_end: a dictionary containing the final model conditions (to be used when working with intervals). It contains:realizations: a np.ndarray of size (n_replay,n_model_equations) containing for eachn_replaythe final state of the model.n_model_equationsis different depending on the blueprint.
insulin_bolus: a dictionary containing the administered insulin boluses during the replay simulation (U/min). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of administered insulin boluses
correction_bolus: a dictionary containing the administered corrective insulin boluses during the replay simulation (U/min). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of administered corrective insulin boluses
insulin_basal: a dictionary containing the administered basal insulin during the replay simulation (U/min). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of administered basal insulin
cho: a dictionary containing the meal intakes had during the replay simulation (g/min). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of meal intakes
hypotreatments: a dictionary containing the hypotreatment intakes had during the replay simulation (g/min). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of hypotreatment intakes
meal_announcment: a dictionary containing the announced CHO intakes had during the replay simulation (g). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of announced CHO intakes
vo2: a dictionary containing the input exercise VO2 used during the replay simulation (-) (NOT YET IMPLEMENTED). It contains:realizations: a np.ndarray of size (n_replay,simulation_length) containing then_replaysimulated series of exercise VO2
sensors: a list ofSensorsobjects of size (n_replay) (to be used when working with intervals).rbg_data: the givendatabut in a proprietaryReplayBGDataformat (to be used for debugging).model: the model used for simulation (to be used for debugging).
Tips
Beside returning the above dictionary, if the save_workspace is set to True, ReplayBG will also save it in a .pkl file using save_name and save_suffix parameters to save it as: results/workspaces/<save_name>_<save_suffix>.pkl.
Replaying single portions of data
To replay single portions of data, i.e., a single meal event or a single day, you can follow this example, which shows how to use a digital twin previously created using the MCMC twinning method (stored in results/mcmc/example.pkl). In this example, we will just simulate a reduction of 20% of the original insulin boluses:
import os
from py_replay_bg.py_replay_bg import ReplayBG
# Set verbosity
verbose = True
plot_mode = False
# Set other parameters for twinning
blueprint = 'multi-meal'
save_folder = os.path.join(os.path.abspath(''))
# Set bw
bw = ... # Set bw
# Instantiate ReplayBG
rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
yts=5, exercise=False,
seed=1,
verbose=verbose, plot_mode=plot_mode)
# Load data and set save_name
data = ...
save_name = 'example'
# Reduce the original boluses by 20%
data.bolus = data.bolus * 0.8
# Replay the twin using the modified data
replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
n_replay=10,
twinning_method='mcmc',
save_workspace=True,
save_suffix='reduced')The results will be saved in <save_folder>/results/workspaces/example_reduced.pkl.
A fully working example can be found in example/code/replay_mcmc.py.
Tips
To do the same but using a digital twin created with the MAP twinning method, just set twinning_method to 'map'. A full example can be found in example/code/replay_map.py.
Replaying portions of data spanning more than one day (i.e., intervals)
To replay portions of data than span multiple days, i.e., an intervals, you need to practically do the following steps:
- initialize empty initial conditions, i.e.,
x0,previous_data_name,sensors. - replay the first day
- extract from the results
x0,previous_data_name,sensorsto be used for the next day - replay the subsequent day 5repeat steps 3 and 4, for each subsequent day of the interval.
To implement these steps you can follow this example, which shows how to replay an interval spanning two days using two digital twins previously created using the MCMC twinning method (stored in results/mcmc/example_1.pkl and results/mcmc/example_2.pkl). In this example, we will just simulate the original insulin boluses:
import os
from py_replay_bg.py_replay_bg import ReplayBG
# Set verbosity
verbose = True
plot_mode = False
# Set other parameters for twinning
blueprint = 'multi-meal'
save_folder = os.path.join(os.path.abspath(''))
# Set bw
bw = ...
x0 = None
previous_data_name = None
sensors = None
# Initialize the list of results
replay_results_interval = []
data_interval = []
# Instantiate ReplayBG
rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
yts=5, exercise=False,
seed=1,
verbose=verbose, plot_mode=plot_mode)
# Set interval to twin
start_day = 1
end_day = 2
# Twin the interval
for day in range(start_day, end_day+1):
# Load data and set save_name
data = ...
save_name = 'example' + str(day)
# Replay the twin with the same input data
replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
twinning_method='mcmc',
n_replay=100,
save_workspace=True,
x0=x0, previous_data_name=previous_data_name, sensors=sensors,
save_suffix='_intervals')
# Append results
replay_results_interval.append(replay_results)
data_interval.append(data)
# Set initial conditions for next day equal to the "ending conditions" of the current day
x0 = replay_results['x_end']['realizations'][0].tolist()
# Set sensors to use the same sensors during the next portion of data
sensors = replay_results['sensors']
# Set previous_data_name
previous_data_name = save_nameThe results will be saved in <save_folder>/results/workspaces/example_1_intervals.pkl and <save_folder>/results/workspaces/example_2_intervals.pkl.
A fully working example can be found in example/code/replay_intervals_mcmc.py.
Tips
To do the same but using a digital twin created with the MAP twinning method, just set twinning_method to 'map'. A full example can be found in example/code/replay_intervals_map.py.
Event handlers
The possibility to alter "offline" the original data before calling rbg.replay() alone is not sufficient for testing, for example, a specific bolus calculation strategy, as the meal/insulin inputs usually depend on the current glucose value and other surrounding conditions which cannot be known before simulation.
In this context, the ReplayBG rbg.replay() method can be fed by a set of callable Python functions, hereafter referred to as "handlers", that will handle the real-time generation of specific events during the simulation.
As such, event handlers allow users to decide whether to use the meal/insulin inputs defined offline by the data parameter of rbg.replay() or those generated online, during simulation, by custom algorithms.
Implemented handlers and logicw
The following scheme defines the logic followed by ReplayBG to decide the input source to use during simulation. Yellow boxes represent input parameters of rbg.replay(), green boxes represent the input data actually used during simulation.
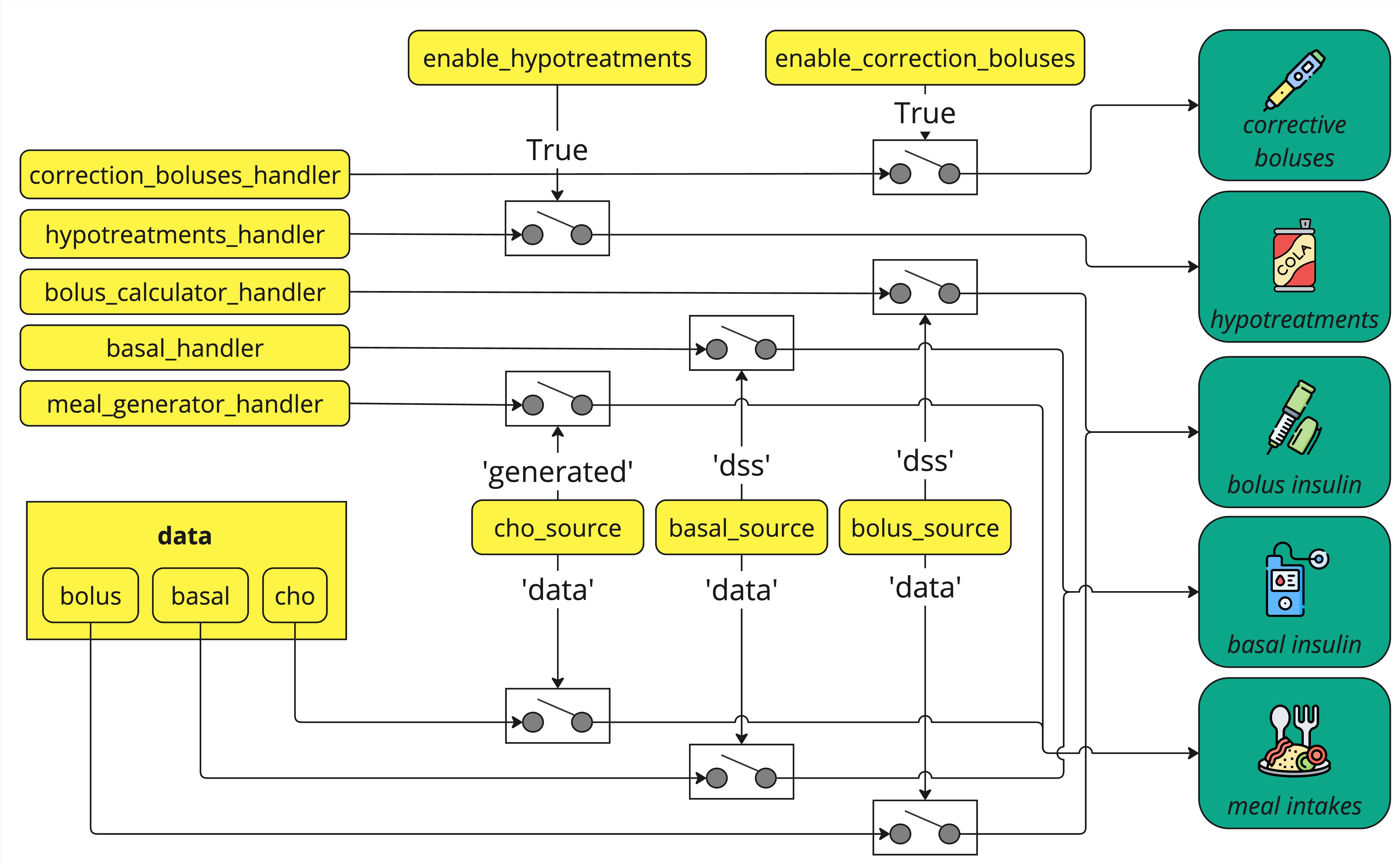
As showed in the figure, five different handlers can be defined:
meal_generator_handler: a function that will replace the carbohydrates ofdata.chowith meal data generated according to a custom user-defined policy whencho_source='generated'.bolus_calculator_handler: a function that will replace the insulin ofdata.boluswith insulin bolus data generated according to a custom user-defined policy whenbolus_source='dss'.basal_handler: a function that will replace the insulin ofdata.basalwith basal insulin data generated according to a custom user-defined policy whenbasal_source='dss'.correction_boluses_handler: a function that will add correction insulin boluses generated according to a custom user-defined policy whenenable_correction_boluses=True.hypotreatments_handler: a function that will add rescue carbs generated according to a custom user-defined policy whenenable_hypotreatments=True.
Each handler must comply to mandatory input/output specifications described in details in the following subsections.
Meal generators
A meal generator handler must be a Python function with 4 outputs and 11 inputs defined as:
def <handler_name>(
glucose: np.ndarray,
meal: np.ndarray,
meal_type: np.ndarray,
meal_announcement: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object,
is_single_meal: bool
) -> tuple[float, float, str, object]:Input parameters
glucose: An np.ndarray as long assimulation_lengthcontaining all the simulated glucose concentrations (mg/dl) up totime_index. The values aftertime_indexshould be ignored by the user.meal: An np.ndarray as long assimulation_lengthcontaining all the meals (g) up totime_index. The values aftertime_indexshould be ignored.meal_type: An np.ndarray as long assimulation_lengthcontaining strings that represent the type of each meal inmeal_announcement:- If blueprint is
single-meal, labels can be:M: main mealO: other meal
- If blueprint is
multi-meal, labels can be:B: breakfastL: lunchD: dinnerS: snackH: hypotreatment The values aftertime_indexshould be ignored.
- If blueprint is
meal_announcement: An np.ndarray as long assimulation_lengthcontaining all the meal announcements (g) up totime_index. The values aftertime_indexshould be ignored.hypotreatments: An np.ndarray as long assimulation_lengthcontaining all the hypotreatment intakes (g/min) up totime_index. If the blueprint is single meal,hypotreatmentswill contain only the hypotreatments generated by this function during the simulation. If the blueprint is multi-meal, hypotreatments will ALSO contain the hypotreatments already present in the givendata(ifcho_source='data') labeled as such. The values aftertime_indexshould be ignored.bolus: An np.ndarray as long assimulation_lengthcontaining all the insulin boluses (U/min) up totime_index. The values aftertime_indexshould be ignored.basal: An np.ndarray as long assimulation_lengthcontaining all the insulin basal (U/min) up totime_index. The values aftertime_indexshould be ignored.time: An np.ndarray as long assimulation_lengthcontaining the time corresponding to the current step (hours) up totime_index. The values aftertime_indexshould be ignored.time_index: The index corresponding to the previous simulation step of the replay simulation. This basically represent the progress of the simulation.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.is_single_meal: A boolean flag indicating if the handler is being used by a single meal blueprint or not.
Output parameters
c: A float that is the meal amount to have at the next simulation step in (g/min), i.e.,time[time_index+1].ma: A float that is the meal announcement (which can be different fromcdue to carb counting errors) to have at the next simulation step in (g/min), i.e.,time[time_index+1].type: A string that is the type of the generated meal. Can be 'M' or 'O' if the blueprint is single meal, 'B' 'L' 'D' or 'S' otherwise.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Default handler
The default bolus calculator handler (which is called when no custom handlers are provided by the user and bolus_source='dss') is defined in py_replay_bg.dss.default_dss_handler and implements a super simple generator that is "put a main meal of 50 g of CHO in the first instant and announce only 40g. If is_single_meal=True, the meal type will be 'M', otherwise it will be a breakfast, 'B'":
def default_meal_generator_handler(
glucose: np.ndarray,
meal: np.ndarray,
meal_type: np.ndarray,
meal_announcement: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object,
is_single_meal: bool
) -> tuple[float, float, str, object]:
# Default output values
c = 0
ma = 0
m_type = ''
# If this is the first time instant...
if time_index == 0:
# ...generate a snack meal of 50 g and announce just 40 g
c = 50
ma = 40
if is_single_meal:
m_type = 'M'
else:
m_type = 'B'
return c, ma, m_type, dssBolus calculators
A bolus calculator handler must be a Python function with 2 outputs and 9 inputs defined as:
def <handler_name>(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:Input parameters
glucose: An np.ndarray as long assimulation_lengthcontaining all the simulated glucose concentrations (mg/dl) up totime_index. The values aftertime_indexshould be ignored by the user.meal_announcement: An np.ndarray as long assimulation_lengthcontaining all the meal announcements (g) up totime_index. The values aftertime_indexshould be ignored.meal_type: An np.ndarray as long assimulation_lengthcontaining strings that represent the type of each meal inmeal_announcement:- If blueprint is
single-meal, labels can be:M: main mealO: other meal
- If blueprint is
multi-meal, labels can be:B: breakfastL: lunchD: dinnerS: snackH: hypotreatment The values aftertime_indexshould be ignored.
- If blueprint is
hypotreatments: An np.ndarray as long assimulation_lengthcontaining all the hypotreatment intakes (g/min) up totime_index. If the blueprint is single meal,hypotreatmentswill contain only the hypotreatments generated by this function during the simulation. If the blueprint is multi-meal, hypotreatments will ALSO contain the hypotreatments already present in the givendata(ifcho_source='data') labeled as such. The values aftertime_indexshould be ignored.bolus: An np.ndarray as long assimulation_lengthcontaining all the insulin boluses (U/min) up totime_index. The values aftertime_indexshould be ignored.basal: An np.ndarray as long assimulation_lengthcontaining all the insulin basal (U/min) up totime_index. The values aftertime_indexshould be ignored.time: An np.ndarray as long assimulation_lengthcontaining the time corresponding to the current step (hours) up totime_index. The values aftertime_indexshould be ignored.time_index: The index corresponding to the previous simulation step of the replay simulation. This basically represent the progress of the simulation.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Output parameters
b: A float that is the bolus insulin to administer at the next simulation step in (U/min), i.e.,time[time_index+1].dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Default handler
The default bolus calculator handler (which is called when no custom handlers are provided by the user and bolus_source='dss') is defined in py_replay_bg.dss.default_dss_handler and implements the standard bolus calculator formula
def standard_bolus_calculator_handler(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:
b = 0
# If a meal is announced...
if meal_announcement[time_index] > 0:
# compute iob
ts = 5
k1 = 0.0173
k2 = 0.0116
k3 = 6.73
iob_6h_curve = np.zeros(shape=(360,))
for t in range(0, 360):
iob_6h_curve[t] = 1 - 0.75 * ((- k3 / (k2 * (k1 - k2)) * (np.exp(-k2 * t / 0.75) - 1) + k3 / (
k1 * (k1 - k2)) * (np.exp(-k1 * t / 0.75) - 1)) / 2.4947e4)
iob_6h_curve = iob_6h_curve[ts::ts]
iob = np.convolve(bolus, iob_6h_curve)
iob = iob[bolus.shape[0] - 1]
# get params
cr = dss.bolus_calculator_handler_params['cr'] if 'cr' in dss.bolus_calculator_handler_params else 10
cf = dss.bolus_calculator_handler_params['cf'] if 'cf' in dss.bolus_calculator_handler_params else 40
gt = dss.bolus_calculator_handler_params['gt'] if 'gt' in dss.bolus_calculator_handler_params else 120
# ...give a bolus
b = np.max([0, meal_announcement[time_index] / cr + (glucose[time_index] - gt) / cf - iob])
return b, dssBasal controllers
A basal controller handler must be a Python function with 2 outputs and 9 inputs defined as:
def <handler_name>(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:Input parameters
glucose: An np.ndarray as long assimulation_lengthcontaining all the simulated glucose concentrations (mg/dl) up totime_index. The values aftertime_indexshould be ignored by the user.meal_announcement: An np.ndarray as long assimulation_lengthcontaining all the meal announcements (g) up totime_index. The values aftertime_indexshould be ignored.meal_type: An np.ndarray as long assimulation_lengthcontaining strings that represent the type of each meal inmeal_announcement:- If blueprint is
single-meal, labels can be:M: main mealO: other meal
- If blueprint is
multi-meal, labels can be:B: breakfastL: lunchD: dinnerS: snackH: hypotreatment The values aftertime_indexshould be ignored.
- If blueprint is
hypotreatments: An np.ndarray as long assimulation_lengthcontaining all the hypotreatment intakes (g/min) up totime_index. If the blueprint is single meal,hypotreatmentswill contain only the hypotreatments generated by this function during the simulation. If the blueprint is multi-meal, hypotreatments will ALSO contain the hypotreatments already present in the givendata(ifcho_source='data') labeled as such. The values aftertime_indexshould be ignored.bolus: An np.ndarray as long assimulation_lengthcontaining all the insulin boluses (U/min) up totime_index. The values aftertime_indexshould be ignored.basal: An np.ndarray as long assimulation_lengthcontaining all the insulin basal (U/min) up totime_index. The values aftertime_indexshould be ignored.time: An np.ndarray as long assimulation_lengthcontaining the time corresponding to the current step (hours) up totime_index. The values aftertime_indexshould be ignored.time_index: The index corresponding to the previous simulation step of the replay simulation. This basically represent the progress of the simulation.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Output parameters
b: A float that is the basal insulin to administer at the next simulation step in (U/min), i.e.,time[time_index+1].dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Default handler
The default basal controler handler (which is called when no custom handlers are provided by the user and basal_source='dss') is defined in py_replay_bg.dss.default_dss_handler and implements a naive algorithm, that is "if glucose < 70, basal = 0; otherwise, basal = 0.01 U/min"
def default_basal_handler(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:
b = 0.01
# If G < 70...
if glucose[time_index] < 70:
# ...set basal rate to 0
b = 0
return b, dssCorrective insulin bolus generators
A corrective insulin bolus generator handler must be a Python function with 2 outputs and 9 inputs defined as:
def <handler_name>(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:Input parameters
glucose: An np.ndarray as long assimulation_lengthcontaining all the simulated glucose concentrations (mg/dl) up totime_index. The values aftertime_indexshould be ignored by the user.meal_announcement: An np.ndarray as long assimulation_lengthcontaining all the meal announcements (g) up totime_index. The values aftertime_indexshould be ignored.meal_type: An np.ndarray as long assimulation_lengthcontaining strings that represent the type of each meal inmeal_announcement:- If blueprint is
single-meal, labels can be:M: main mealO: other meal
- If blueprint is
multi-meal, labels can be:B: breakfastL: lunchD: dinnerS: snackH: hypotreatment The values aftertime_indexshould be ignored.
- If blueprint is
hypotreatments: An np.ndarray as long assimulation_lengthcontaining all the hypotreatment intakes (g/min) up totime_index. If the blueprint is single meal,hypotreatmentswill contain only the hypotreatments generated by this function during the simulation. If the blueprint is multi-meal, hypotreatments will ALSO contain the hypotreatments already present in the givendata(ifcho_source='data') labeled as such. The values aftertime_indexshould be ignored.bolus: An np.ndarray as long assimulation_lengthcontaining all the insulin boluses (U/min) up totime_index. The values aftertime_indexshould be ignored.basal: An np.ndarray as long assimulation_lengthcontaining all the insulin basal (U/min) up totime_index. The values aftertime_indexshould be ignored.time: An np.ndarray as long assimulation_lengthcontaining the time corresponding to the current step (hours) up totime_index. The values aftertime_indexshould be ignored.time_index: The index corresponding to the previous simulation step of the replay simulation. This basically represent the progress of the simulation.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Output parameters
cb: A float that is the correction bolus to administer at the next simulation step in (U/min), i.e.,time[time_index+1].dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Default handler
The default corrective insulin bolus generator handler (which is called when no custom handlers are provided by the user and enable_correction_boluses=True) is defined in py_replay_bg.dss.default_dss_handler and implements a naive algorithm, that is "take a correction bolus of 1 U every 1 hour while above 250 mg/dl"
def corrects_above_250_handler(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:
cb = 0
# If glucose is higher than 250...
if glucose[time_index] > 250:
# ...and if there are no boluses in the last 60 minutes, then take a CB
if time_index >= 60 and not np.any(bolus[(time_index - 60):time_index]):
cb = 1
return cb, dssHypotreatment generators
A hypotreatment generator handler must be a Python function with 2 outputs and 9 inputs defined as:
def <handler_name>(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:Input parameters
glucose: An np.ndarray as long assimulation_lengthcontaining all the simulated glucose concentrations (mg/dl) up totime_index. The values aftertime_indexshould be ignored by the user.meal_announcement: An np.ndarray as long assimulation_lengthcontaining all the meal announcements (g) up totime_index. The values aftertime_indexshould be ignored.meal_type: An np.ndarray as long assimulation_lengthcontaining strings that represent the type of each meal inmeal_announcement:- If blueprint is
single-meal, labels can be:M: main mealO: other meal
- If blueprint is
multi-meal, labels can be:B: breakfastL: lunchD: dinnerS: snackH: hypotreatment The values aftertime_indexshould be ignored.
- If blueprint is
hypotreatments: An np.ndarray as long assimulation_lengthcontaining all the hypotreatment intakes (g/min) up totime_index. If the blueprint is single meal,hypotreatmentswill contain only the hypotreatments generated by this function during the simulation. If the blueprint is multi-meal, hypotreatments will ALSO contain the hypotreatments already present in the givendata(ifcho_source='data') labeled as such. The values aftertime_indexshould be ignored.bolus: An np.ndarray as long assimulation_lengthcontaining all the insulin boluses (U/min) up totime_index. The values aftertime_indexshould be ignored.basal: An np.ndarray as long assimulation_lengthcontaining all the insulin basal (U/min) up totime_index. The values aftertime_indexshould be ignored.time: An np.ndarray as long assimulation_lengthcontaining the time corresponding to the current step (hours) up totime_index. The values aftertime_indexshould be ignored.time_index: The index corresponding to the previous simulation step of the replay simulation. This basically represent the progress of the simulation.dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Output parameters
ht: A float that is the hypotreatment to administer at the next simulation step in (g), i.e.,time[time_index+1].dss: An object of typeDSS. Might contain parameters usable by the handler. More aboutdssis discussed below in the Thedssparameter section.
Default handler
The default hypotreatment generator handler (which is called when no custom handlers are provided by the user and enable_hypotreatments=True) is defined in py_replay_bg.dss.default_dss_handler and implements the ADA guidelines for hypotreatments generation that are ""take a hypotreatment of 10 g every 15 minutes while in hypoglycemia"
def ada_hypotreatments_handler(
glucose: np.ndarray,
meal_announcement: np.ndarray,
meal_type: np.ndarray,
hypotreatments: np.ndarray,
bolus: np.ndarray,
basal: np.ndarray,
time: np.ndarray,
time_index: int,
dss: object
) -> tuple[float, object]:
ht = 0
# If glucose is lower than 70...
if glucose[time_index] < 70:
# ...and if there are no CHO intakes in the last 15 minutes, then take an HT
if time_index >= 15 and not np.any(hypotreatments[(time_index - 15):time_index]):
ht = 15
return ht, dssThe dss parameter
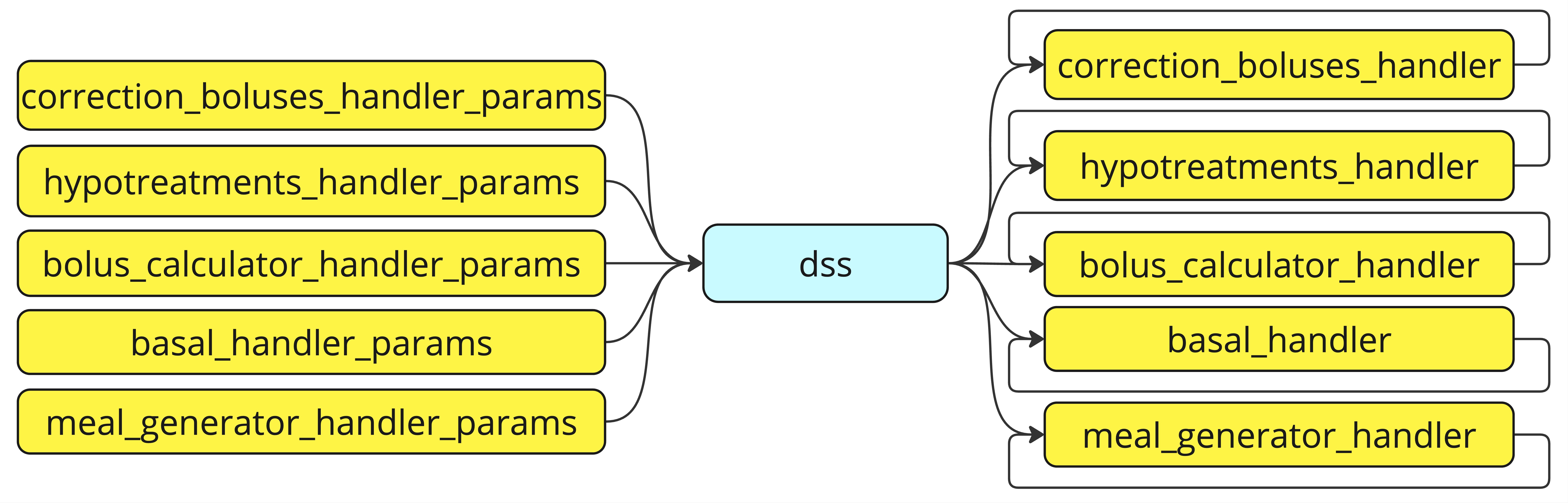
Each of the handler has the dss parameter as input/output.
dss is an object of type DSS, created within the rbg.replay() method, that, as schematized in figure above, has two main goals:
- it acts as a container of any hyperparameter and/or data one wants to pass to the custom handler functions.
- it serves as memory area
Regarding the first point, the dss objects will contain the correction_boluses_handler_params, hypotreatments_handler_params, bolus_calculator_handler_params, basal_handler_params, and meal_generator_handler_params dictionaries given in input to rbg.replay(). As such, if one needs to pass specific parameters to the custom handler, he/she should provide them in the dictionaries as subsequently access them as dss.correction_boluses_handler_params, dss.hypotreatments_handler_params, dss.bolus_calculator_handler_params, dss.basal_handler_params, and dss.meal_generator_handler_params, respectively.
Regarding the second point, being dictionaries mutable in Python, if needed it is possible to store values inside such parameters of dss so that the handler will be able to access to them in the next call of the function.