Choosing Blueprint
Choosing Blueprint
Choosing the right blueprint for the digital twin is one of the key choices to be made when using ReplayBG. This must be done when a ReplayBG object needs to be created.
As a matter of fact, the blueprint, which is associated to a specific mathematical model, will practically represent the resulting digital twin and will define its domain of applicability as well as what are its final "capabilities".
So, what are the possibilities? And, most importantly, how to decide?
There are two possibilities:
blueprint='single-meal': by "single meal" one can refer to a specific period of time when a specific subject had only 1 main meal and a corresponding insulin basal-bolus administration. Usually, this period of time spans maximum 6/8 hours, starts near such main meal, and ends just before the subsequent main meal and/or after a reasonable amount of time.blueprint='multi-meal': by "multi meal" one can refer to a specific period of time when a specific subject had more than 1 main meal and a corresponding insulin basal-bolus administration regimen. One can think to such period of time by thinking to a day, when multiple meals occur, or even multiple days.
As such, the driver through the choice of the right blueprint are the data to be used to create the digital twin. Anyway, ReplayBG will not choose automatically the right blueprint based on the input data, so this aspect is up to the user.
In the following, details on the two blueprint structures are reported, as well as their key capabilities and constraints.
Tips
Generally, the multi meal blueprint is the most common choice since usually one deals with data that include more than 1 meal and more than 6/8 hours.
Single-meal
Usage
To use the single-meal blueprint, set the blueprint parameter to single-meal when building the ReplayBG object:
rbg = ReplayBG(blueprint='single-meal', ...)More details can be found in The ReplayBG Object page.
Structure
The single-meal blueprint is composed of four main subsystems: subcutaneous insulin absorption, oral glucose absorption, glucose-insulin kinetics, CGM sensor error.
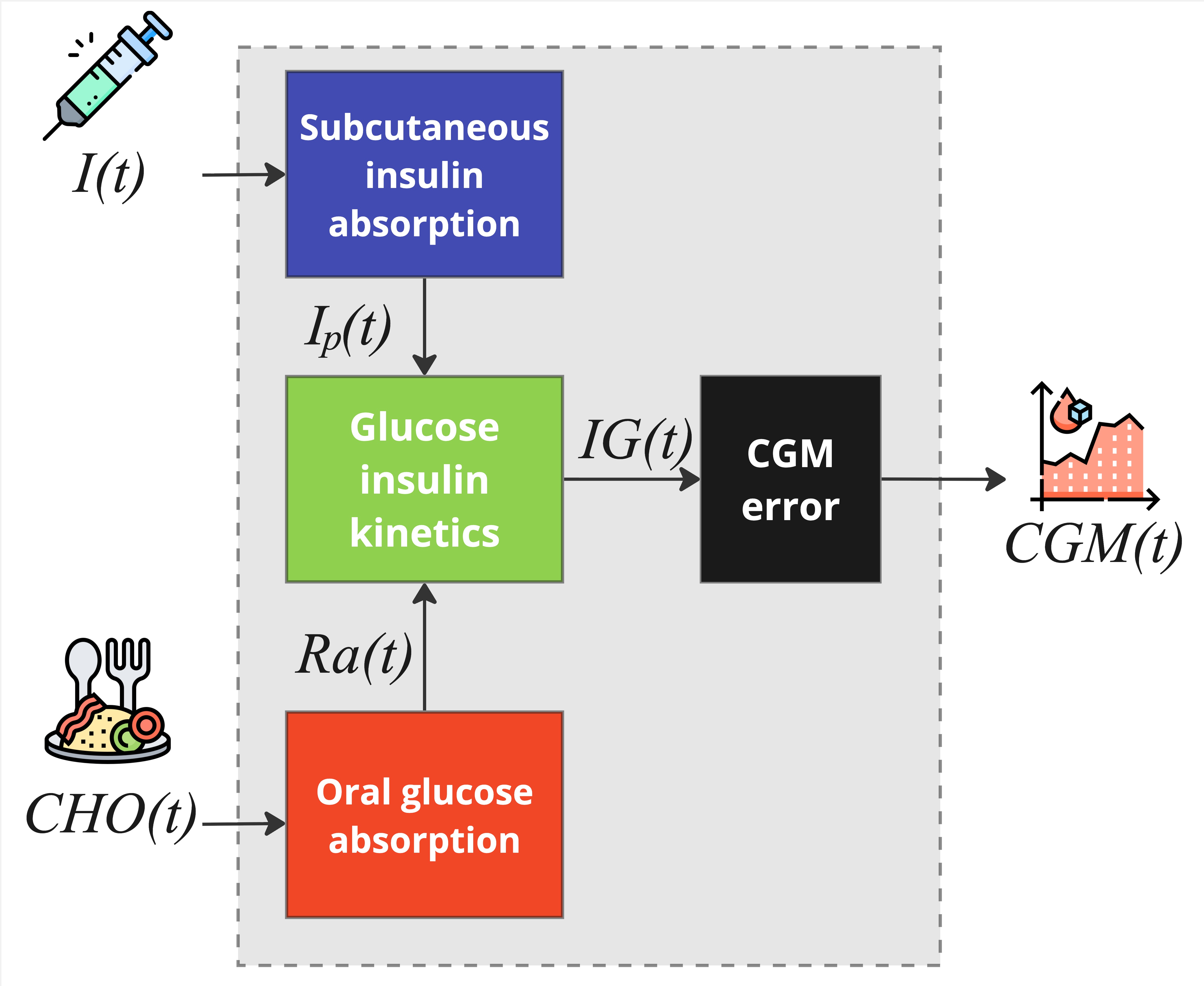
Subcutaneous Insulin Absorption Subsystem
The subsystem of subcutaneous insulin absorption system is composed of three compartments and describes the absorption dynamics of exogenous insulin infusion to the plasma. Exogenous insulin
where
Oral Glucose Absorption Subsystem
The subsystem of oral glucose absorption system describes the gastro-intestinal tract as a three-compartment system: the first two compartments quantify the glucose in the stomach, while the third compartment models the upper small intestine where CHO is absorbed. Model equations are:
where
where
Glucose-Insulin Kinetics Subsystem
The subsystem is composed of three compartments, the first describing the effect of insulin action and oral glucose rate of appearance on plasma glucose concentration, the second quantifies the impact of plasma insulin concentration on insulin action, and the last one represents the transport of glucose from plasma to the interstitium. Model equations are:
where
where
CGM sensor error model
The subsystem describes the CGM sensor error model. For more details on the CGM error model, please refer to the CGM Error Model page.
Overall model
The resulting model of T1D glucoregulatory system described above, can be rewritten as:
where
What can I do?
Using the single-meal blueprint you can create digital twin using portion of data that contains just one main meal, e.g., a breakfast, a lunch, a dinner, or a snack, and spans no more than 8 hours.
This is because its equations can just represent one single meal absorption (i.e., via
Furthermore, since the equations start for steady-state conditions additional data requirements must be taken into account. Particularly, user must be careful in selecting a data starting point that is sufficiently far, e.g., 4 hours, from the last recorded insulin/meal input, thus guaranteeing with a certain degree of confidence that "data are starting from steady-state" as well.
More details and guidance on such requirements, as well as on the format of input data, please refer to the Data Requirements page.
Multi-meal
Usage
To use the multi-meal blueprint, set the blueprint parameter to multi-meal when building the ReplayBG object:
rbg = ReplayBG(blueprint='multi-meal', ...)More details can be found in The ReplayBG object page.
Structure
The multi-meal blueprint is an expanded version of the single-meal one. It is composed of four main subsystems: subcutaneous insulin absorption, oral glucose absorption, glucose-insulin kinetics, CGM sensor error.
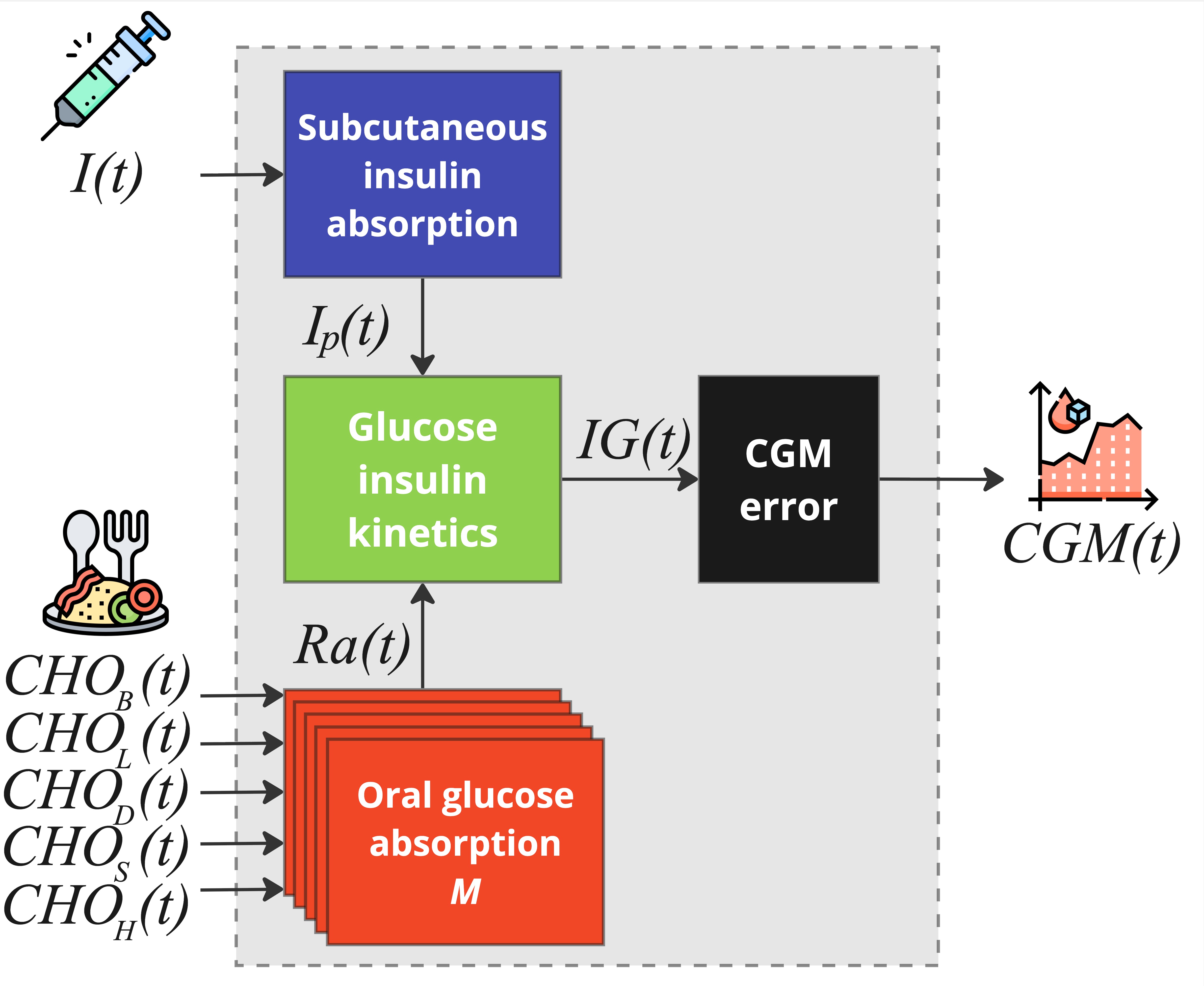
Subcutaneous Insulin Absorption Subsystem
The subsystem of subcutaneous insulin absorption coincides with the one of the single-meal blueprint. It is composed of three compartments and describes the absorption dynamics of exogenous insulin infusion to the plasma. Exogenous insulin
where
Oral Glucose Absorption Subsystem
The subsystem of the oral glucose absorption of the single-meal blueprint, provides an accurate description of oral glucose absorption rate and dynamics for a single meal intake, but it is not suitable when multiple different meals are present. For this reason, it has been replaced with a "multi-stomach" system, where each "stomach" is dedicated to describe one of the specific meal absorption dynamics that can be observed through a single-day:
where suffix
It is important to note that, since multiple snacks and/or hypoglycemia treatments can occur in a single day, single absorption rate constants,
Glucose-Insulin Kinetics Subsystem
The subsystem is composed of three compartments, the first describing the effect of insulin action and oral glucose rate of appearance on plasma glucose concentration, the second quantifies the impact of plasma insulin concentration on insulin action, and the last one represents the transport of glucose from plasma to the interstitium. Model equations are:
where
where
CGM sensor error
The subsystem describes the CGM sensor error model. For more details on the CGM error model, please refer to the CGM Error Model page.
Overall model
The resulting model of T1D glucoregulatory system described above, can be rewritten as:
where
What can I do?
Using the multi-meal blueprint you can create digital twin using portion of data that span a single day.
This is because its equations can just represent the meal absorption of a set of meal intakes usually observed throughout a single day, i.e, a breakfast, a lunch, and so on, via
Furthermore, since the equations start for steady-state conditions additional data requirements must be taken into account. Particularly, user must be careful in selecting a data starting point that is sufficiently far, e.g., 4 hours, from the last recorded insulin/meal input, thus guaranteeing with a certain degree of confidence that "data are starting from steady-state" as well.
More details and guidance on such requirements, as well as on the format of input data, please refer to the Data Requirements page.
So, what if you want to use portions of data that span more than one day? The solution is to create multiple digital twins, each representing a single day, and then "glue" them together.
Practically, this translates in the following steps:
- splitting the data into single days
- creating the digital twin corresponding to the first day by starting from steady state conditions
- creating the digital twins of the second day using the same twinning procedure but setting the initial model conditions to the final model conditions of the first day.
- iterating, for each subsequent day of the interval, step 3.
This process eliminates the need for the initial steady-state assumption for all days except the first, enabling the twinning and simulation of longer, more comprehensive data intervals.
More details and guidance on how to create digital twins using just a single day or more than one, please refer to the Twinning Procedure page.