Twinning Procedure
Twinning Procedure
The twinning procedure represents the key step to create a digital twin. It consists of personalizing the blueprint of choice (more details about blueprints in Choosing Blueprint page), and practically, given
where
How to twin?
The twinning procedure in ReplayBG can be performed using the twin method of the ReplayBG object, which is formally defined as:
rbg.twin(data: pd.DataFrame, bw: float, save_name: str,
twinning_method: str = 'mcmc',
extended: bool = False, find_start_guess_first: bool = False,
n_steps: int = 50000, save_chains: bool = False,
u2ss: float | None = None, x0: np.ndarray | None = None, previous_data_name: str | None = None,
parallelize: bool = False, n_processes: int | None = None,
) -> NoneInput parameters
data: A Pandas dataframe which contains the data to be used by the tool. For more information ondataformat and requirements see Data Requirements page.bw: A float representing the patient's body weight in kg.save_name: A string used to label, thus identify, each output file and result.u2ss, optional, default:None: A float representing the steady state of the basal insulin infusion (e.g., the average basal insulin) in mU/(kg*min). IfNone, it will be set to the average of the basal insulin in input.x0, optional, default:None: An np.ndarray, containing the initial model conditions. IfNone, the model will start to the default steady state.previous_data_name, optional, default:None: A string representing the name of the previous data portion. This is used to correcly "transfer" the initial model conditions to the current portion of data. Practically, this is equal to thesave_nameused during the creation of the digital twin related to the previous portion of data. It must be set ifx0is notNone.twinning_method, optional,{'mcmc', 'map'}, default:'mcmc': A string used to select the method to be used to twin the model.extended, optional, default :False: A flag indicating whether to use "extended" portions of data for twinning. For more information see below.find_start_guess_first: optional, default :False: A flag indicating whether to set the start parameter guess by running a round of MAP before twinning.n_steps, optional, default:50000: An integer representing the number of steps to use by the'mcmc'procedure. This is ignored iftwinning_methodis'map'.save_chains, optional, default :False: A boolean that specifies whether to save additional results of the mcmc twinning method. . This is ignored iftwinning_methodis'map'.parallelize, optional, default:False: A boolean that specifies whether to parallelize the twinning process. This is strongly advised, but it is up to the user.n_processes, optional, default:None: An integer defining the number of processes to be spawn ifparallelizeisTrue. IfNone, the whole number of CPU cores is used.
More on save_name parameter
The save_name parameter is of paramount importance. It will be used to "label" the resulting digital twin and save the resulting parameters in the results/ folder.
Specifically, for a given twinning_method of choice, the digital twin will be saved in a file results/<twinning_method>/<twinning_method>_<save_name>.pkl.
More on data used for twinning
If you have questions like:
- What happens if I'm using the multi-meal blueprint and I use a day worth of data that do not include any lunch event?
- What happens if I use the multi-meal blueprint and I use data that span just half a day?
- ...
The answer is simple: parameters "associated" to events that are not present in the data used to twin will be just set to pouplation values. This means, for example, that in the first case,
Twinning single portions of data
To twin single portions of data, i.e., a single meal event or a single day, you can follow this example, which shows how to create a digital twin using the MCMC twinning method:
import os
from py_replay_bg.py_replay_bg import ReplayBG
# Set verbosity
verbose = True
plot_mode = False
# Set the number of steps for MCMC
n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
# Set other parameters for twinning
blueprint = 'multi-meal'
save_folder = os.path.join(os.path.abspath(''))
parallelize = True
# Set bw and u2ss
bw = ... # set bw
u2ss = ... # set u2ss
# Instantiate ReplayBG
rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
yts=5, exercise=False,
seed=1,
verbose=verbose, plot_mode=plot_mode)
# Load data and set save_name
data = ... # load your data
save_name = 'example' # set a save_name
print("Twinning " + save_name)
# Run twinning procedure
rbg.twin(data=data, bw=bw, save_name=save_name,
twinning_method='mcmc',
parallelize=parallelize,
n_steps=n_steps,
u2ss=u2ss)The resulting digital twin will be saved in <save_folder>/results/mcmc/mcmc_example.pkl.
The fully working example can be found in example/code/twin_mcmc.py.
Tips
To do the same but using the MAP twinning method, just set twinning_method to 'map'. The full example can be found in example/code/twin_map.py.
Twinning portions of data spanning more than one day (i.e., intervals)
To twin portions of data than span multiple days, i.e., an intervals, you need to create multiple digital twins, each representing a single day, and then "glue" them together.
Practically, this translates in the following steps:
- splitting the data into single days
- creating the digital twin corresponding to the first day by starting from steady state conditions
- creating the digital twins of the second day using the same twinning procedure but setting the initial model conditions to the final model conditions of the first day.
- iterating, for each subsequent day of the interval, step 3.
To implement these steps you can follow this example, which shows how to create the twins using the MCMC twinning method:
import os
from py_replay_bg.py_replay_bg import ReplayBG
# Set verbosity
verbose = True
plot_mode = False
# Set the number of steps for MCMC
n_steps = 5000 # 5k is for testing. In production, this should be >= 50k
# Set other parameters for twinning
blueprint = 'multi-meal'
save_folder = os.path.join(os.path.abspath(''))
parallelize = True
# Set bw and u2ss
bw = ... # set bw
u2ss = ... # set u2ss
x0 = None # At the beginning the model will start from steady-state, so x0 must be None
previous_data_name = None # Same for previous_data_name
# Instantiate ReplayBG
rbg = ReplayBG(blueprint=blueprint, save_folder=save_folder,
yts=5, exercise=False,
seed=1,
verbose=verbose, plot_mode=plot_mode)
# Set interval to twin
start_day = 1
end_day = 2
# Twin the interval
for day in range(start_day, end_day+1):
# Load data and set save_name
data = ... # load your data
save_name = 'example' + str(day) # set a save_name
# Run twinning procedure
rbg.twin(data=data, bw=bw, save_name=save_name,
twinning_method='mcmc',
parallelize=parallelize,
n_steps=n_steps,
x0=x0, u2ss=u2ss, previous_data_name=previous_data_name)
# Replay the twin with the same input data
replay_results = rbg.replay(data=data, bw=bw, save_name=save_name,
twinning_method='mcmc',
save_workspace=True,
x0=x0, previous_data_name=previous_data_name,
save_suffix='_twin_intervals_mcmc')
# Set initial conditions for next day equal to the "ending conditions" of the current day
x0 = replay_results['x_end']['realizations'][0].tolist()
# Set previous_data_name
previous_data_name = save_nameThe resulting digital twins will be saved in results/mcmc/mcmc_example_1.pkl and results/mcmc/mcmc_example_2.pkl.
The fully working example can be found in example/code/twin_intervals_mcmc.py.
Basically, the main differences with the case of single portions of data are:
- you need to set and manage
previous_data_name. For the first day, it isNone, then it must be set tosave_name - you need to set and manage
x0. For the first day, it isNone, then it must be set to the final conditions obtained by running a replay with the same input data, i.e.,x0 = replay_results['x_end']['realizations'][0].tolist()
Tips
To do the same but using the MAP twinning method, just set twinning_method to 'map'. The full example can be found in example/code/twin_intervals_map.py.
Warning
When working with intervals, you should use the same u2ss otherwise the digital twins will have different steady-state conditions and equilibrium across the interval (which does not make sense).
Twinning using extended portions of data
When twinning, it might be useful to run the twinning procedure using more data than actually necessary.
This is particularly useful to avoid that wrong parameters are set for the last part of the data, e.g., at dinner time. This is also reffered to as "tail effect".
As an example, think about having a dinner event at 22:00 and the portion of data you are using ends at 23:00. In this scenario, the twinning procedure will just have 1 hour worth of data to "understand" what is a plausible value for the dinner's absorption rate, which might be challenging.
For this reason, if one has enough data after the actual portion of data to be used for twinning, it is possible to set the extended parameter to True and also leverage those data points for improving the process.
Warning
This feature is implemented for the multi-meal blueprint only.
Practically speaking, one has to follow three steps.
Step 1. Prepare the input data appropriately
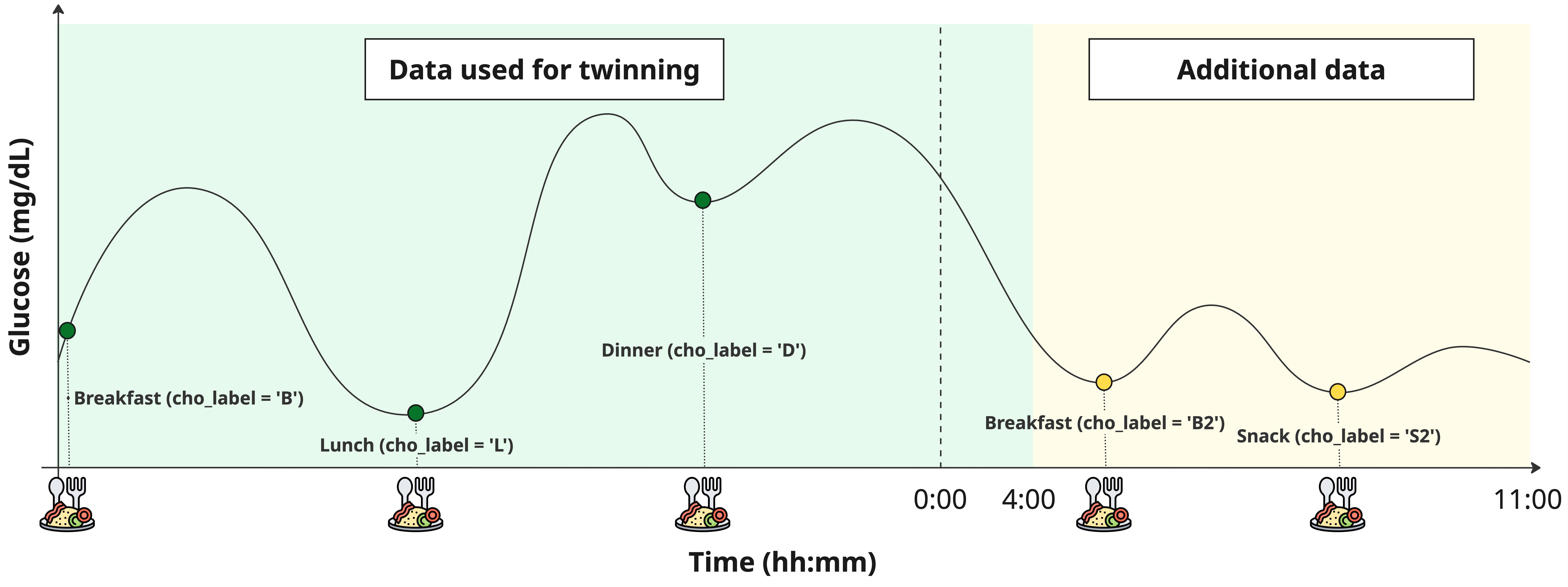
As shown in the figure, data must be prepared before running the twinning procedure if extended=True.
To do that, the user must simply flag the meal events of the additional portion of data adding a 2, i.e., B -> B2, L -> L2, S -> S2. No need to flag also the insulin boluses. Also, no need to flag differently hypotreatment events.
Tips
Since the insulin sensitivity
Tips
To avoid to increase the computation time too much, the additional portion of data should not go > 11:00 of the next day.
An example code for running this procedure is:
# Get the hours
hours = np.array([t.hour for t in data.t])
# Find the last idx of the first portion of data < 4:00
idx_split = np.where(hours == 3)[0][-1] + 1
# Find the idxs with the labels to be modified and modify them
idx_b2 = np.where(data.cho_label.values == 'B')[0]
idx_b2 = idx_b2[idx_b2 > idx_split]
idx_l2 = np.where(data.cho_label.values == 'L')[0]
idx_l2 = idx_l2[idx_l2 > idx_split]
idx_s2 = np.where(data.cho_label.values == 'S')[0]
idx_s2 = idx_s2[idx_s2 > idx_split]
data.loc[idx_b2, "cho_label"] = 'B2'
data.loc[idx_l2, "cho_label"] = 'L2'
data.loc[idx_s2, "cho_label"] = 'S2'Step 2. Set extended parameter
The second step must simply set extended equal to True and run rbg.twin(), e.g.:
rbg.twin(...,
extended=True)Step 3. Run replays
Finally, remember to remove the additional portion of data before replaying, e.g.:
# Cut data up to idx_split
data = data.iloc[0:idx_split, :]
# Replay the twin
replay_results = rbg.replay(...)Tips
Example code snippets can be found in example/code/twin_mcmc_extended.py and example/code/twin_map_extended.py. An example of already prepared data is stored in example/data/data_day_1_extended.csv.
MCMC
Theoretical flavours
The twinning procedure 'mcmc' of ReplayBG personalizes
Briefly, utilizing the a priori information
where
This chain is used to represent
From the implementative point-of-view, the current version of ReplayBG adopts the affine invariant ensemble sampler for MCMC (AIES-MCMC) proposed by Goodman and Weare in 2010 implemented in the state-of-the-art emcee package. This method offers several advantages over traditional MCMC sampling methods and demonstrates excellent performance in the literature for multidimensional, generic model parameter distributions. A comprehensive discussion of AIES-MCMC is beyond the scope of this documentation, and the interested readers are referred to Goodman and Weare for further details.
What will be saved
The resulting results/mcmc/mcmc_<save_name>.pkl file contains a Python dictionary with the following fields:
draws: a dictionary with the following fields:Gb: a dictionary containing the estimated values of theGbparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
SG: a dictionary containing the estimated values of theSGparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
ka2: a dictionary containing the estimated values of theka2parameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kd: a dictionary containing the estimated values of thekdparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kempt: a dictionary containing the estimated values of thekemptparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
SI(only ifblueprint='single-meal'): a dictionary containing the estimated values of theSIparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
SI_B(only ifblueprint='multi-meal', and ifdatacontains data within the 4 AM - 11 AM time range): a dictionary containing the estimated values of theSI_Bparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
SI_L(only ifblueprint='multi-meal', and ifdatacontains data within the 11 AM - 5 PM time range): a dictionary containing the estimated values of theSI_Lparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
SI_D(only ifblueprint='multi-meal', and ifdatacontains data within the 5 PM - 4 AM time range): a dictionary containing the estimated values of theSI_Dparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs(only ifblueprint='single-meal'): a dictionary containing the estimated values of thekabsparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs_B(only ifblueprint='multi-meal', and ifdatacontains a meal breakfast eventB): a dictionary containing the estimated values of thekabs_Bparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs_L(only ifblueprint='multi-meal', and ifdatacontains a meal lunch eventL): a dictionary containing the estimated values of thekabs_Lparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs_D(only ifblueprint='multi-meal', and ifdatacontains a meal dinner eventD): a dictionary containing the estimated values of thekabs_Dparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs_S(only ifblueprint='multi-meal', and ifdatacontains a meal snack eventS): a dictionary containing the estimated values of thekabs_Sparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
kabs_H(only ifblueprint='multi-meal', and ifdatacontains a meal hypotreatment eventH): a dictionary containing the estimated values of thekabs_Hparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
beta(only ifblueprint='single-meal'): a dictionary containing the estimated values of thebetaparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
beta_B(only ifblueprint='multi-meal', and ifdatacontains a meal breakfast eventB): a dictionary containing the estimated values of thebeta_Bparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
beta_L(only ifblueprint='multi-meal', and ifdatacontains a meal lunch eventL): a dictionary containing the estimated values of thebeta_Lparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
beta_D(only ifblueprint='multi-meal', and ifdatacontains a meal dinner eventD): a dictionary containing the estimated values of thebeta_Dparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
beta_S(only ifblueprint='multi-meal', and ifdatacontains a meal snack eventS): a dictionary containing the estimated values of thebeta_Sparameter in four different sampled forms:samples_1000: a list with 1000 realizationssamples_100: a list with 100 realizationssamples_10: a list with 10 realizationssamples_1: a list with just 1 realization
u2ss: the value ofu2ssused during twinningsampler(only ifsave_chain=True): the MCMC sampler objecttau(only ifsave_chain=True): the value of the estimated autocorrelation timethin(only ifsave_chain=True): the MCMC thinning factorburnin(only ifsave_chain=True): the number of burn-in samples
Tips
Since hypotreatments are usually related to fast absorbing meals, beta_H is not estimated and fixed to 0.
Tips
To select between the sampled forms samples_1000, samples_100, samples_10, samples_1, use the n_replay parameter of the replay method of a ReplayBG object and set it to 1000, 100, 10, or 1, accordingly. See the Replaying page for more details.
Tips
To ensure to have the same resulting confidence interval no matter the chosen sampled form, the values contained in sampled_100 are chosen as those corresponding to the 1, 2, ..., 100 percentiles of the confidence interval obtained by simulating the 1000 glucose profiles using sampled_1000. Similarly, the values contained in sampled_10 are chosen as those corresponding to the 1, 10, ..., 100 percentiles of the confidence interval obtained by simulating the 1000 glucose profiles using sampled_1000. Finally, the values of samples_1 are chosen as those corresponding to the median glucose profile of the confidence interval obtained using sampled_1000.
MAP
Theoretical flavours
The twinning procedure 'map' of ReplayBG personalizes
Briefly, utilizing the a priori information
where
The result will be a value of
Warning
Conversely to MCMC, the procedure will produce just one value of
From the implementative point-of-view, the current version of ReplayBG adopts the modified Powell algorithm implemented the minimize function of scipy.optimize.
Tips
The MAP twinning method is lot faster than MCMC, however, it is supposed to be less robust to local minima. As such, for more accurate digital twins, MCMC is advised, while for prototyping, MAP is a more than valuable choice.
What will be saved
The resulting results/map/map_<save_name>.pkl file will contain the following:
The resulting results/<twinning_method>/<twinning_method>_<save_name>.pkl file contains a Python dictionary with the following fields:
draws: a dictionary with the following fields:Gb: the estimated values of theGbparameterSG: the estimated values of theSGparameterka2: the estimated values of theka2parameterkd: the estimated values of thekdparameterkempt: the estimated values of thekemptparameterSI(only ifblueprint='single-meal'): the estimated values of theSIparameterSI_B(only ifblueprint='multi-meal', and ifdatacontains data within the 4 AM - 11 AM time range): the estimated values of theSI_BparameterSI_L(only ifblueprint='multi-meal', and ifdatacontains data within the 11 AM - 5 PM time range): the estimated values of theSI_LparameterSI_D(only ifblueprint='multi-meal', and ifdatacontains data within the 5 PM - 4 AM time range): the estimated values of theSI_Dparameterkabs(only ifblueprint='single-meal'): the estimated values of thekabsparameterkabs_B(only ifblueprint='multi-meal', and ifdatacontains a meal breakfast eventB): the estimated values of thekabs_Bparameterkabs_L(only ifblueprint='multi-meal', and ifdatacontains a meal lunch eventL): the estimated values of thekabs_Lparameterkabs_D(only ifblueprint='multi-meal', and ifdatacontains a meal dinner eventD): the estimated values of thekabs_Dparameterkabs_S(only ifblueprint='multi-meal', and ifdatacontains a meal snack eventS): the estimated values of thekabs_Sparameterkabs_H(only ifblueprint='multi-meal', and ifdatacontains a meal hypotreatment eventH): the estimated values of thekabs_Hparameterbeta(only ifblueprint='single-meal'): a dictionary containing the estimated values of thebetaparameterbeta_B(only ifblueprint='multi-meal', and ifdatacontains a meal breakfast eventB):- the estimated values of the
beta_Bparameter beta_L(only ifblueprint='multi-meal', and ifdatacontains a meal lunch eventL): the estimated values of thebeta_Lparameterbeta_D(only ifblueprint='multi-meal', and ifdatacontains a meal dinner eventD): the estimated values of thebeta_Dparameterbeta_S(only ifblueprint='multi-meal', and ifdatacontains a meal snack eventS): the estimated values of thebeta_Sparameter
u2ss: the value ofu2ssused during twinning
Tips
Since MAP has no sampled forms, the n_replay parameter of the replay method of a ReplayBG object is ignored. See the Replaying page for more details.